LTRpred(ict): de novo annotation of young and intact retrotransposons 
Transposable elements (TEs) comprise vast parts of eukaryotic genomes. In the past, TEs were seen as selfish mobile elements capable of populating a host genome to increase their chances for survival. By doing so they leave traces of junk DNA in host genomes that are usually regarded as by-products when sequencing, assembling, and annotating new genomes.
However, this picture is slowly changing (Drost & Sanchez, 2019) and TEs have been shown to be involved in generating a diverse range of novel phenotypes.
Today, the de novo detection of transposable elements is performed by annotation tools which try to detect any type of repeated sequence, TE family, or remnand DNA loci that can be associated with a known transposable element within a genome assembly. The main goal of such efforts is to retrieve a maximum amount of loci that can be associated with TEs. If successful, such annotation can then be used to mask host genomes and to perform classic (phylo-)genomics studies focusing on host genes.
More than 600 repeat and TE annotation tools have been developed so far. Most of them are designed and optimized to annotate either the entire repeat space or specific superfamilies of TEs and their DNA remnants.
The LTRpred pipeline has a different goal than all other annotation tools. It focuses particularly on LTR retrotransposons and aims to annotate only functional and potentially mobile elements. Such type of annotation is crucial for studying retrotransposon activity in eukaryotic genomes and to understand whether specific retrotransposon families can be activated artificially and harnessed to mutagenize genomes at much faster speed.
In detail, LTRpred will take any genome assembly file in fasta format as input and will generate a detailed annotation of functional and potentially mobile LTR retrotransposons.
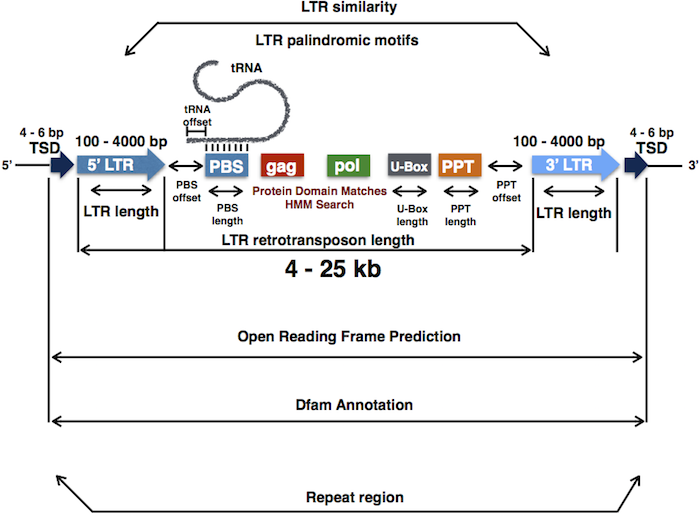
Users can consult a comprehensive Introduction to the LTRpred pipeline to get familiar with the tool.
Install
The fastest way to install LTRpred is via a Docker container. Please make sure to read the detailed installation instructions to be able to pass data to the container.
# retrieve docker image from dockerhub
docker pull drostlab/ltrpred
# run ltrpred container
docker run --rm -ti drostlab/ltrpred
# start R prompt within ltrpred container
~:/app# RUsers who wish to run the LTRpred Docker container in a conda environment can use the following approach based on UDocker (Many thanks to Ilja Bezrukov).
Accessing LTRpred Container via RStudio
A more interactive way of performing analyses with LTRpred is via the RStudio version of LTRpred. In this LTRpred Docker Container users can access LTRpred within the container via Rstudio.
# retrieve docker image from dockerhub
docker pull drostlab/ltrpred_rstudio
# run ltrpred container
docker run -e PASSWORD=ltrpred --rm -p 8787:8787 -ti drostlab/ltrpred_rstudioTo open RStudio and interact with the container go to your standard web browser and type in the following URL:
http://localhost:8787
Username: rstudio
Password: ltrpredUsers can choose a custom password if they wish.
Within RStudio you can now run the example:
LTRpred::LTRpred(genome.file = system.file("Hsapiens_ChrY.fa", package = "LTRpred"))Users can exit the container by pressing Ctrl + c multiple times.
Please find all details here about how to use the Rstudio version here.
Citation
Please cite the following paper when using LTRpred for your own research:
HG Drost. LTRpred: de novo annotation of intact retrotransposons. Journal of Open Source Software, 5(50), 2170 (2020).
Tutorials
Quick Start
The fastest way to generate a LTR retrotransposon prediction for a genome of interest (after installing all prerequisite command line tools) is to use the LTRpred() function and relying on the default parameters. In the following example, a LTR transposon prediction is performed for parts of the Human Y chromosome.
# Perform de novo LTR transposon prediction for the Human Y chromosome
LTRpred::LTRpred(genome.file = system.file("Hsapiens_ChrY.fa", package = "LTRpred"))When running your own genome, please specify genome.file = "path/to/your/genome.fasta instead of system.file(..., package = "LTRpred"). The command system.file(..., package = "LTRpred") merely references the path to the example file stored in the LTRpred package itself.
This tutorial introduces users to LTRpred:
Users can also read the tutorials within (RStudio) :
library(LTRpred)
browseVignettes("LTRpred")You can also find a list of all available LTRpred functions here: https://hajkd.github.io/LTRpred/reference/index.html
Studies that successfully used LTRpred to annotate functional retrotransposons
Z Wang & D Baulcombe. Transposon age and non-CG methylation. Nature Communications, 11, 1221 (2020).
JH Collins, KW Keating, TR Jones, S Balaji, CB Marsan et al. Engineered yeast genomes accurately assembled from pure and mixed samples. Nature communications, 12, 1485 (2021).
H Kundariya et al. MSH1-induced heritable enhanced growth vigor through grafting is associated with the RdDM pathway in plants Nature Communications, 11, 5343 (2020).
J Cho, M Benoit, M Catoni, HG Drost, A Brestovitsky, M Oosterbeek and J Paszkowski. Sensitive detection of pre-integration intermediates of LTR retrotransposons in crop plants. Nature Plants, 5, 26-33 (2019).
M Benoit, HG Drost, M Catoni, Q Gouil, S Lopez-Gomollon, DC Baulcombe, J Paszkowski. Environmental and epigenetic regulation of Rider retrotransposons in tomato. PloS Genetics, 15(9): e1008370 (2019).
Nguinkal et al. The First Highly Contiguous Genome Assembly of Pikeperch (Sander lucioperca), an Emerging Aquaculture Species in Europe Genes, 0(9), 708 (2019).
E Cerruti, C Gisbert, HG Drost, D Valentino, E Portis, L Barchi, J Prohens, S Lanteri, C Comino, M Catoni. Epigenetic bases of grafting-induced vigour in eggplant. bioaRxiv (2019).
P Gan, R Hiroyama, A Tsushima, S Masuda et al. Subtelomeric regions and a repeat-rich chromosome harbor multicopy effector gene clusters with variable conservation in multiple plant pathogenic Colletotrichum species bioRxiv (2020)
J Wang et al. Gigantic Genomes Can Provide Empirical Tests of TE Dynamics Models–An Example from Amphibians. bioRxiv, (2020).
Y Ayukawa et al. A pair of effectors encoded on a conditionally dispensable chromosome of Fusarium oxysporum suppress host-specific immunity. bioRxiv, (2020).
C Meguerditchian, A Ergun, V Decroocq, M Lefebvre et al. Pipeline to detect the relationship between transposable elements and adjacent genes in host genome bioRxiv, (2021).